You are not logged in. (Sign In)
Hippocampal subfield measurements for the early diagnosis AD and prediction of cognitive decline
PI: Lei Wang
Summary:
Dr. Wang is working with Drs. Michael Weiner/Susanne Mueller (co-PIs, UCSF lead site), Dr. Paul Yushkevich (U Penn), and Dr. Koen van Leemput (Harvard) on a project entitled "ADNI 2 add-on project: Superior power of hippocampal subfield measurements compared to conventional hippocampal volumetry for the early diagnosis AD and prediction of AD related cognitive decline." This project is funded by the Alzheimer Association, aiming to use multiple approach to measure hippocampal subfields using a new high-resolution T2-weighted sequence to detect the earliest hippocampal manifestations of AD.
This cross-sectional study will test if subfield volumetry is indeed superior to standard hippocampal volumetry to identify the earliest manifestation of Alzheimer’s disease.
Funding:
- Alzheimer Association: ADNI 2 add on project: Hippocampal Subfield Volumetry (PI: NU Subaward - Lei Wang (co-PIs Michael Weiner/Susanne Mueller, UCSF)) - ADNI 2 add-on project: Superior power of hippocampal subfield measurements compared to conventional hippocampal volumetry for the early diagnosis AD and prediction of AD related cognitive decline - Northwestern Subaward. The project is a three year cross-sectional multi-site collaborative study between Penn Image Computing and Science Lab (PICSL, University of Pennsylvania), Athinoula A Martinos Center for Biomedical Imaging (AAMCBI, Harvard), Neuroimaging and Applied Computational Anatomy Lab (NIACAL, Northwestern University) and the Center for Imaging of Neurodegenerative Diseases (CIND, University of California, San Francisco)). Dr Weiner at CIND will be responsible for the administrative oversight of the project. Dr S. Mueller at CIND will be responsible for the scientific oversight of the entire project. PICSL, AAMBI and NIACAL will be affiliated research sites with one co-investigator each. The overall goal of the project is to test a. if subfield volumetry is superior to conventional hippocampal volumetry to detect the earliest manifestation of beginning AD and b. to identify which of the currently available subfield volumetry approaches has the highest sensibility and sensitivity to differentiate between healthy controls and MCI and between amyloid positive and negative subjects. (07/01/2012 - 06/30/2015).
Details:
The overall goal of this cross-sectional study is to test, if subfield volumetry is indeed superior to standard hippocampal volumetry to identify the earliest manifestation of Alzheimer’s disease. For the purpose of this application, the earliest manifestation of AD is defined as either normal cognitive function with evidence for pathological CNS amyloid deposition (HCbeta+ as determined by PET imaging with a amyloid specific tracer) and/or as mild cognitive impairment without dementia with (MCIbeta+) or without (MCIbeta-) pathological CNS amyloid deposition. 200 Subjects (100 MCI, 100 HC) fulfilling these criteria and amyloid negative controls (HCbeta-) will be selected from the study population currently recruited for Alzheimer’s Disease Neuroimaging Initiative(ADNI) 2. ADNI 2 is the continuation of ADNI 1 and ADNI GO and will enroll 150 healthy controls and 250 MCI (100 early and 150 late) who will all undergo F18 amyloid PET with florpiramine and MR imaging on 3T magnets. If it is assumed that the population recruited for ADNI 2 is similar to the population recruited for ADNI1 it can be expected that about 30- 50% of the cognitively intact subjects and about 75 % of the MCI subjects will have pathological CNS amyloid levels. The ADNI 2 MR protocol includes two (1 non-accelerated, 1 accelerated) 3D T1 whole brain images at each timepoint (required for Methods A,B ,C) but no high resolution T2 weighted sequence (required for Method D). It is therefore planned to identify a number of x suitable ADNI sites who will add this particular sequence to the standard ADNI 2 imaging protocol. Subfield measurements will be obtained using methods A-D (manual marking will be performed in a subset of 100 (50 controls, 50 MCI) subjects). All T1 images acquired for ADNI 2 are routinely processed with Freesurfer and so the unedited hippocampus volumes provided by Freesurfer which has shown an excellent performance in ADNI1 will be used to address the question if subfield volumetry is indeed superior to total hippocampal volumetry to detect the earliest hippocampal manifestations of AD. Several histopathological studies have shown that the earliest manifestation of AD in the hippocampus affect the dorsal region of the CA1 sector (5-7). Based on that observation, the a priori hypotheses will focus on volume changes in CA1. Specifically the following hypotheses will be tested:
Specific Aim 1. Amyloid positive subjects (HCbeta+ and MCIbeta+) have a smaller CA1 than amyloid negative subjects (HCbeta-, MCIbeta-). Hypothesis 1a: MCIbeta+ subjects have reduced CA1 volumes compared to MCIbeta-, HCbeta+ and HCbeta-. The CA1 volume differences will be largest between MCIbeta+ and HCbeta- and smallest between MCIbeta+ and MCIbeta-. Hypothesis 1b. HCbeta+ have smaller CA1 volumes than HCbeta-.
Specific Aim 2. Subfield volumetry is superior to standard hippocampal volumetry (unedited hippocampal volume provided by Freesurfer) for the detection of localized (CA1) volume losses. Hypothesis 2 a. Subfield volumetry will have a greater power to detect small CA1 volume differences (HCbeta+ vs. HCbeta-, MCIbeta+ vs MCIbeta- and HCbeta+) than standard hippocampal volumetry. Hypothesis 2b. Subfield volumetry approaches using internal hippocampal landmarks for subfield labeling (Methods C,D) have greater power to detect small CA1 volume differences than subfield volumetry approaches based on shape deformation (Methods A,B).
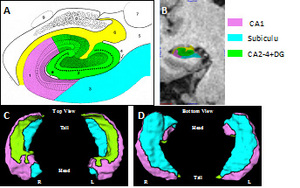
Image Processing: Method A: The two (accelerated, non-acclerated) T1-weighted MRI from each subject will be aligned and averaged to obtain a low-noise, high-contrast image. A fully automated Freesurfer + Large Deformation Diffeomorphic Metric Mapping (FSLDDMM) pipeline (12) will be used to generate the whole hippocampal surface in each target scan from an existing atlas. In the atlas, boundaries between hippocampal subfields have been previously identified on its surface according to underlying hippocampal image subvolumes, creating the CA1, Sub and combined CA2-4+DG subfield zones (13,14). An example of the atlas hippocampal subfields is illustrated in Figure 1. The FreeSurfer image analysis suite provides a noisy labeling of the hippocampus, which will be used to initially align the hippocampal region of interest, followed by LDDMM where neuroanatomical targets (i.e., whole hippocampal surface) are generated from the atlas via a flow of diffeomorphisms with the smoothness of brain structure boundaries preserved (20). Following FSLDDMM, deformation vector fields between each subject’s whole hippocampal surface and a reference surface (e.g., global average) will first be computed. The atlas subfield boundaries are then transferred onto each individual hippocampal surface via the above diffeomorphic transformations to define vector fields for each anatomical subfield (i.e., CA 1, Sub, CA2-4+DG). Finally, vector analysis methods such as principal components analysis will be used to compare subfield shape between groups.